Approach
The internal exposure to chemical agents may contribute significantly to chronic health impacts. The IP Exposome undertakes targeted interdisciplinary research into molecular mechanisms of toxicity through the combined application of toxicogenome (transcriptome, proteome, metabolome, epigenome), exposome (including the adductome), and bioinformatics platforms (e. g. MetaPro). Phenotype-level anchoring of the high-dimensional omics profiles to classical toxicological endpoints will facilitate the biological interpretation. To this end, we build on established collaborations between chemists, biochemists, molecular and systems biologists, toxicologists and ecotoxicologists, human and environmental epidemiologists, and computational scientists.
Two complementary strategies have been envisaged to reach the goal of unravelling exposome signatures related to Adverse Outcome Pathways and associated chronic health impacts:
- Analysis of exposome and omics patterns of bio-specimens from diseased and healthy individuals (top-down approach)
- Analysis of omics, exposome, and phenotype responses resulting from controlled single and multiple exposure in vitro (bottom-up or case study approach)
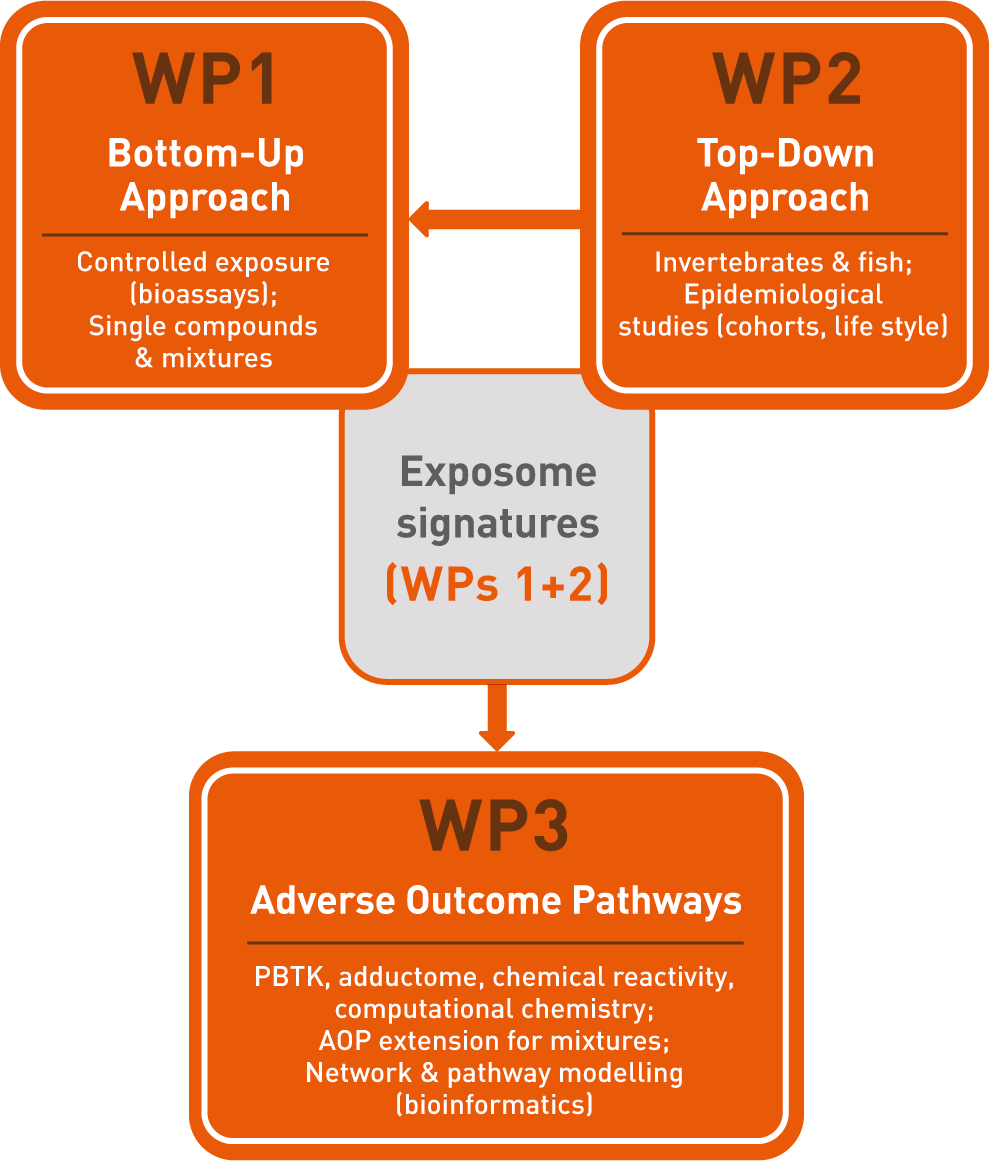
The three work packages (WP) of IP "Exposome"
For the top-down approach, bio-specimens from the prospective epidemiological mother-child study LiNA (lifestyle and environmental factors and their influence on new-borns' allergy risk) will be subjected to omics and exposome investigations, to identify further model compounds for the case-study programme. The LiNA study focuses on immune system-derived diseases, mainly lung inflammation and allergy. Depending on intermediate project results, biospecimens from additional cohorts focussing on other diseases will be included in the investigation.
For the bottom-up approach, model compounds and priority pollutants representing known modes of action are tested alone and in combinations in diverse immortal cell models, reporter gene assays and human T lymphocytes, focussing on metabolic dysfunctions and immune system impairments. Moreover, zebrafish embryos are employed, providing an in vitro link between human and environmental health. Candidates for model compounds to be selected include electrophiles, oxidative stress inducers, and receptor-binding compounds that have been identified as key components of the exposome. Following the AOP concept as mentioned above, special emphasis is devoted to unravelling the points of departure that discriminate between reversible perturbations that can be accommodated by the biological system (adaptive stress response), and exposome signatures characterised further through associated omics profiles that result in biological damage.
The work package structure
IP Exposome is comprised of three work packages. The rationale for this structure is to comprehensively address the cross-cutting character of adverse outcome pathways leading from molecular initiating events to chronic diseases and adverse outcomes in the environment, and the challenges in relating internal dose signatures to external exposure. The three WPs are interlinked through methods such as the omics platform and advanced analytics, through the major types of physiological dysfunctions under analysis, and through an overall common set of perturbing factors of interest.
WP 1: Bottom-Up Approach
WP 2: Top-Down Approach
WP 3: Adverse Outcome Pathways
